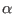 -carbon
-carbon
- STRUCTURES
- acc
- acc
- acceptance ratio
- ADD_BOND
| ADD_BEND
| ADD_TORS
| PRINT
- ADD_TORS
- ADD_TORS
- ADD_BEND
- ADD_BEND
- ADD_BOND
- ADD_BOND
- ADD_STR_COM
- ADD_STR_BENDS
| ADD_STR_COM
- ADD_STR_BONDS
- ADD_STR_BONDS
- ADD_STR_TORS
- ADD_STR_TORS
- adding a bending
- ADD_STR_BONDS
- adding a COM-COM stretching
- ADD_STR_BENDS
- adding a harmonic distance constraint
- &POTENTIAL
- adding an harmonic torsion
- ADD_STR_COM
- ADD_TPG SOLUTE
- ADD_TPG SOLUTE
- ADD_UNITS
- ADD_UNITS
- ADJUST_BONDS
- ADJUST_BONDS
- AGBNP
- MINIMIZE
- Alchemical transformations
- Implementation in ORAC
| CUTOFF
| STEER_PATH
- definition of the alchemical portion of the solute
- DEFINE_ALCHEMICAL_ATOM
- printout of the work done
- PLOT
- mass scaling
- SCALE_MASS
- alkanes
- Applications
- AMBER force field
- Subdivision of the ``Bonded''
- &ANALYSIS
- &ANALYSIS
- ANDERSEN
- ANDERSEN
- Andersen, H.C.
- ANDERSEN
- angular_cutoff
- HBONDS
- angular cutoff
- HBONDS
- animation
- using ORAC generated file
- PLOT
- animation from xyz file
- DEF_FRAGMENT
- ANNEALING
- UPDATE
| ANNEALING
- DCD
- ASCII
| ASCII_OUTBOX
| DCD
- asymmetric unit
- SPACE_GROUP
- atom_record
- DUMP
- atomic charges
- subtracting excess charge
- DEF_SOLUTE
- atomic scaling
- Equivalence of Atomic and
- Liouvillean split for
- Liouvillean Split and Multiple
- atoms
- atoms
- AUTO_DIHEDRAL
- AUTO_DIHEDRAL
- average
- GOFR
- averaged
- STRUCTURES
- B-spline interpolation
- The smooth particle mesh
- backbone
- STRUCTURES
| backbone
- writing the coordinates of [] atoms
- PLOT
- barostat
- The Parrinello-Rahman-Nosé Extended Lagrangian
- BATTERIES
- replica exchange method
- BATTERIES
- BENDING
- BENDING
- printing out
- PRINT_TOPOLOGY
- bending potential
- Subdivision of the ``Bonded''
| DEBUG
- BENDINGS
- BENDING
- Bennett acceptance ratio
- Calculating Ensemble Averages Using
| Determination of the potential
- Berendsen H.
- Equivalence of Atomic and
- Berne B. J
- Multiple time steps integration
- BOND
- BOND
- bonded potential
- Multiple Time Steps Algorithms
- subdivision of
- Multiple Time Steps Algorithms
- bonds
- bonds
- BPTI
- Group Scaling and Molecular
- B-spline
- of the direct lattice potential
- ERFC_SPLINE
- BUSSI
- BUSSI
- Bussi, G.
- BUSSI
- canonical transformations
- Canonical Transformation and Symplectic
- CELL
- CELL
- center of mass
- REPLICATE
- CG
- MINIMIZE
- CHANGE_CELL
- CHANGE_CELL
- CHECK_COORD_OFF
- CHECK_COORD_OFF
- Ciccotti G.
- Equivalence of Atomic and
- cofm
- THERMOS
- COM-COM added bond
- ADD_STR_BENDS
- compiling ORAC
- Serial version
- compute accessibility
- VORONOI
- compute contac_solute
- VORONOI
- compute neighbors
- VORONOI
- compute volume
- VORONOI
- config.h file
- How to set dimensions
| How to set dimensions
- configure file
- How to set dimensions
- conjugate gradient
- MINIMIZE
| THERMOS
- constant pressure
- scaling
- SCALING
- constant pressure simulation
- Multiple Time Steps Algorithms
| ISOSTRESSXY
- constant temperature simulation
- Multiple Time Steps Algorithms
| THERMOS
| THERMOS
- CONSTRAINT
- CONSTRAINT
- constraints
- ADD_STR_TORS
| CHECK_COORD_OFF
- printing out
- PRINT_TOPOLOGY
- with r-RESPA
- Potential Subdivision and Multiple
- CONTROL
- CONTROL
- COORDINATES
- COORDINATES
| COORDINATES
- of the solvent
- ADD_UNITS
| READ_SOLVENT
- Crooks theorem
- Steered Molecular Dynamics
- CRYSTAL
- CRYSTAL
- crystal structure
- READ_PDB
- crystal symmetry
- SPACE_GROUP
- crystal to orthogonal matrix
- READ_CO
- crystallographic parameters
- CHANGE_CELL
- cutoff
- Multiple time steps integration
| The smooth particle mesh
| Subdivision the Non Bonded
| Subdivision the Non Bonded
| Subdivision the Non Bonded
| CUTOFF
| ERFC_SPLINE
| GOFR
| VORONOI
- for hydrogen bonds
- HBONDS
- in the reciprocal lattice
- Electrostatic Corrections for the
- reciprocal lattice
- ERFC_SPLINE
- DCD
- generating a file
- DCD
- DEBUG
- DEBUG
- decaalanine
- Enhanced sampling in atomistic
- defaults
- THERMOS
- DEF_FRAGMENT
- DEF_FRAGMENT
- DEFINE_ALCHEMICAL_ATOM
- DEFINE_ALCHEMICAL_ATOM
- DEF_SOLUTE
- DEF_SOLUTE
- density of states
- Subdivision of the ``Bonded''
- dielectric constant
- Multiple time steps integration
| Electrostatic Corrections for the
| PRINT_DIPOLE
- diffusion
- TIME_CORRELATIONS
- diffusion coefficient
- &PROPERTIES
- dihed
- dihed
- dihedral angle
- Subdivision of the ``Bonded''
- dihedral angle in torsions
- Topology
- dimensions
- changing the, in ORAC
- How to set dimensions
- dipole
- &PROPERTIES
- Direct lattice Coulomb potential
- Soft-core variant for alchemical transformations
- Production of the MD
| SHIFT-POT
- direct lattice potential
- The smooth particle mesh
- direct potential
- subdivision of
- Subdivision the Non Bonded
- dirty
- MTS_RESPA
- discrete time propagator
- Liouville Formalism: a Tool
| Liouville Formalism: a Tool
| Potential Subdivision and Multiple
| Subdivision the Non Bonded
- DIST_FRAGMENT
- DEF_FRAGMENT
| DIST_FRAGMENT
- dist_max
- FREQUENCIES
- dived_step
- TIME_CORRELATIONS
- don
- don
- driven thermal changes
- PLOT
| STEER
- driving external potential
- &POTENTIAL
- DUMP
- DUMP
- dumping the restart file
- RESTART
- DYNAMIC
- DYNAMIC
- dynamical matrix
- FREQUENCIES
- eigenvectors
- FREQUENCIES
| FREQUENCIES
- electrostatic correction
- ERFC_SPLINE
- electrostatic corrections
- Subdivision the Non Bonded
- electrostatic potential
- The smooth particle mesh
- subdivision of
- Subdivision the Non Bonded
- energy equipartition
- THERMOS
- energy_then_die
- MTS_RESPA
- enhanced sampling
- Multiple time steps integration
- equations of motion
- Canonical Transformation and Symplectic
- for Parrinello-Rahman-Nosé Hamiltonian
- The Parrinello-Rahman-Nosé Hamiltonian and
- equilibration
- TIME
- ERF_CORR
- ERF_CORR
- ERFC_SPLINE
- ERFC_SPLINE
- error function
- The smooth particle mesh
| ERFC_SPLINE
- EWALD
- EWALD
- Ewald method
- Multiple time steps integration
| The smooth particle mesh
- electrostatic corrections
- Subdivision the Non Bonded
- in multiple time scales integrators
- MTS_RESPA
- intramolecular correction
- ERF_CORR
- intramolecular self term
- The smooth particle mesh
- self energy
- Electrostatic Corrections for the
- setting work array dimensions
- How to set dimensions
- smooth particle mesh
- The smooth particle mesh
- excess charge
- DEF_SOLUTE
- extended Lagrangian
- Multiple Time Steps Algorithms
- Fast switching Double Annihilation method
- Calculation of the alchemical
- FIX_FREE_ENERGY(&SGE)
- FIX_FREE_ENERGY
- fluctuation theorem
- The Crooks theorem
- force breakup
- Potential Subdivision and Multiple
- force field
- Subdivision of the ``Bonded''
| Input to ORAC : Force
- input parameters from ASCII file
- READ_PRM_ASCII
- input parameters from tpgprm file
- READ_TPGPRM
- force field printout
- DYNAMIC
- FORCE_FIELD
- FORCE_FIELD
- fractional translations
- SPACE_GROUP
- fragment
- writing coordinates of
- PLOT
- free energy
- Enhanced sampling in atomistic
- FREQUENCIES
- FREQUENCIES
- FS-DAM
- Calculation of the alchemical
- fudge factor
- LJ-FUDGE
- generalized Born solvent model
- MINIMIZE
- GENERATE
- GENERATE
- glycine
- REPL_RESIDUE
- GOFR
- GOFR
- group scaling
- The Parrinello-Rahman-Nosé Hamiltonian and
| Equivalence of Atomic and
| Liouvillean Split and Multiple
| SCALING
- Liouvillean split for
- Liouvillean Split and Multiple
- GROUP_CUTOFF
- GROUP_CUTOFF
- Hamilton
- equations
- Canonical Transformation and Symplectic
| Liouville Formalism: a Tool
- harmonic constraints
- &POTENTIAL
| ADD_STR_BONDS
- on the center of mass
- ADD_STR_BENDS
- harmonic frequencies
- FREQUENCIES
- HBONDS
- HBONDS
- heavy_atoms
- VORONOI
- Hermitian operator
- Liouville Formalism: a Tool
- histogram
- HBONDS
- history file
- DUMP
| DUMP
| TRAJECTORY
| &PROPERTIES
- auxiliary file
- DUMP
- H-MASS
- H-MASS
- hydrogen bond
- HBONDS
| acc
- acceptor and donor
- don
- imphd
- imphd
- implicit solvent
- MINIMIZE
- improper torsion
- Subdivision of the ``Bonded''
| PRINT_TOPOLOGY
| DEBUG
| imphd
- definition of, in the parameter file
- TORSION IMPROPER
- &INOUT
- &INOUT
- INSERT
- INSERT
- inst_xrms
- STRUCTURES
- &INTEGRATOR
- &INTEGRATOR
- reversible
- Liouville Formalism: a Tool
- symplectic
- Enhanced sampling in atomistic
| Liouville Formalism: a Tool
- interchange matrix
- SPACE_GROUP
- ISEED
- ISEED
- isokinetic scaling
- SCALE_MASS
- ISOSTRESS
- ISOSTRES
| ISOSTRESSXY
- isothermal-isobaric ensemble
- Applications
- I-TORSION
- I-TORSION
- jacobian
- Canonical Transformation and Symplectic
- Jarzynski identity
- The Crooks theorem
- JOIN
- JOIN
- JORGENSEN
- JORGENSEN
- KEEP_BONDS
- KEEP_BONDS
- k-ewald
- MTS_RESPA
- leap frog algorithm
- Liouville Formalism: a Tool
- Legendre transformation
- The Parrinello-Rahman-Nosé Hamiltonian and
- Lennard-Jones
- Applications
| NONBONDED
- Soft-core variant for alchemical transformations
- Production of the MD
| SHIFT-POT
- cutoff
- Subdivision the Non Bonded
- parameters
- NONBONDED
- linked cell
- LINKED_CELL
| UPDATE
- LINKED_CELL
- LINKED_CELL
- Liouville
- formalism
- Liouville Formalism: a Tool
- Liouvillean
- Multiple time steps integration
| Potential Subdivision and Multiple
| Subdivision the Non Bonded
- split of Parrinello-Rahman-Nosé
- Equivalence of Atomic and
- liquid water
- Electrostatic Corrections for the
- LJ-FUDGE
- LJ-FUDGE
- Lucy's functions
- Implementation in ORAC
- lysozyme
- JOIN
- Markovian process
- Steered Molecular Dynamics
- Martyna G
- Multiple time steps integration
- mass
- isokinetic scaling
- SCALE_MASS
- of the Nosé thermostat
- THERMOS
- specifying the type atomic
- NONBONDED
- maximum likelihood
- Determination of the potential
- MAXRUN
- MAXRUN
- MBAR
- Calculating Ensemble Averages Using
- MDSIM
- MDSIM
- mean square displacement
- TIME_CORRELATIONS
- membrane
- simulation at constant pressure in the NPT ensemble
- ISOSTRES
- memory demand in ORAC
- How to set dimensions
- Message Passing library interface
- Serial version
- &META
- &META
- metadynamics
- Enhanced sampling in atomistic
| Metadynamics Simulation: history-dependent algorithms
| ADD_BOND
| ADD_BEND
| ADD_TORS
- Gaussian and Lucy's function
- Implementation in ORAC
- multiple walkers
- Enhanced sampling in atomistic
- well-tempered metadynamics
- Implementation in ORAC
- minimization
- with dielectric continuum
- MINIMIZE
- MINIMIZE
- MINIMIZE
- mixing rules
- NONBONDED
- molecular scaling
- Liouvillean Split and Multiple
| Group Scaling and Molecular
| SCALING
- Liouvillean split for
- Liouvillean Split and Multiple
- MPI
- Serial version
- MTS_RESPA
- MTS_RESPA
- multiple Bennett acceptance ratio
- Calculating Ensemble Averages Using
- multiple restarts in parallel simulation
- RESTART
- multiple time steps
- Multiple time steps integration
| Switching to Other Ensembles
- for Parrinello-Rahman-Nosé Hamiltonian
- The Parrinello-Rahman-Nosé Hamiltonian and
- neighbor list
- LINKED_CELL
| UPDATE
- for hydrogen bonds
- HBONDS
- setting work arrays dimensions
- How to set dimensions
- non bonded potential
- subdivision of
- The smooth particle mesh
- non-bonded potential
- Multiple Time Steps Algorithms
- Nosé thermostat
- The Parrinello-Rahman-Nosé Extended Lagrangian
| THERMOS
| THERMOS
- no_step
- FREQUENCIES
- NPT ensemble
- Multiple Time Steps Algorithms
| THERMOS
- NPT simulation
- ISOSTRES
- NVT ensemble
- Multiple Time Steps Algorithms
| Multiple Time Steps Algorithms
| Group Scaling and Molecular
| PLOT
| THERMOS
- occupy
- DUMP
- omit_angles
- omit_angles
- OpenMP
- Serial version
- pair correlation function
- GOFR
- PAR prefix
- changing the
- SETUP
- parallel job
- Launching multiple independent simulations
- BATTERIES
- parallel version
- compiling
- Parallel version
- REM algorithm
- SETUP
- steered molecular dynamics simulations
- ADD_STR_BONDS
- &PARAMETERS
- &PARAMETERS
- Parrinello-Rahman-Nosé Extended Lagrangian
- Multiple Time Steps Algorithms
| SCALING
- PARXXXX directories
- SETUP
| &SGE
| Parallel version
- PDB
- READ_PDB
| SOLVENT
- generating a file
- ASCII
| ASCII_OUTBOX
- writing the [] file to disk
- PLOT
- PLOT
- PLOT
- PLOT FRAGMENT
- DEF_FRAGMENT
| DIST_FRAGMENT
- PMF
- Steered Molecular Dynamics
- position Verlet
- Liouville Formalism: a Tool
- &POTENTIAL
- &POTENTIAL
- bending
- Subdivision of the ``Bonded''
- bonded
- Multiple Time Steps Algorithms
- non-bonded
- Multiple Time Steps Algorithms
- of mean force
- Steered Molecular Dynamics
- stretching
- Subdivision of the ``Bonded''
- subdivision of
- Potential Subdivision and Multiple
- potential of mean force
- determination of via the Crooks theorem
- Determination of the potential
- potential subdivision
- Multiple Time Steps Algorithms
| Subdivision the Non Bonded
- for the AMBER force field
- Subdivision of the ``Bonded''
| Subdivision the Non Bonded
- PRESSURE
- parameter in the config.h file
- How to set dimensions
- control for membrane simulation
- ISOSTRES
- simulation with isotropic and anisotropic stress tensor
- Multiple Time Steps Algorithms
| ISOSTRESSXY
- primaDORAC
- READ_TPG_ASCII
- print
- HBONDS
| PRINT
- for harmonic calculations
- FREQUENCIES
- replica exchange method
- PRINT
| PRINT_ENERGY
- PRINT_DIAGNOSTIC
- replica exchange method
- PRINT_DIAGNOSTIC
- PRINT_DIPOLE
- PRINT_DIPOLE
- print_histo
- HBONDS
- printing the force field parameters
- DYNAMIC
| DEBUG
- printing topology information
- DEBUG
- PRINT_WHAM(&SGE)
- PRINT_ACCEPTANCE_RATIO
| PRINT_WHAM
- PRINT_TOPOLOGY
- PRINT_TOPOLOGY
- propagator
- Liouville Formalism: a Tool
- discrete time
- Liouville Formalism: a Tool
- stepwise
- Liouville Formalism: a Tool
- proper torsion
- Subdivision of the ``Bonded''
| PRINT_TOPOLOGY
| DEBUG
- definition of, in the parameter file
- TORSION PROPER
- frequency range
- Subdivision of the ``Bonded''
- &PROPERTIES
- &PROPERTIES
- PROPERTY
- PROPERTY
- protein
- printing out the sequence
- PRINT_TOPOLOGY
- giving the input sequence in ORAC
- JOIN
- p_test
- MTS_RESPA
- QQ-FUDGE
- QQ-FUDGE
- r-RESPA
- Potential Subdivision and Multiple
- energy conservation
- Applications
| Applications
- for NPT ensemble
- Equivalence of Atomic and
- input examples
- MTS_RESPA
- performances
- Applications
| Applications
- use in ORAC
- MTS_RESPA
- with Parrinello-Rahman-Nosé Hamiltonian
- The Parrinello-Rahman-Nosé Hamiltonian and
- radial_cutoff
- HBONDS
- radial distribution function
- &PROPERTIES
- RATE
- RATE
- RATTLE
- Constraints and r-RESPA
- reaction coordinate
- Enhanced sampling in atomistic
| The Crooks theorem
- reaction field
- Multiple time steps integration
- READ
- READ
- READ_CO
- READ_CO
- reading the restart file
- RESTART
| CONTROL
- READ_PDB
- READ_PDB
- READ_PRM_ASCII
- READ_PRM_ASCII
- READ_SOLVENT
- READ_SOLVENT
- READ_TPG_ASCII
- READ_TPG_ASCII
- READ_TPGPRM
- READ_TPGPRM
- reciprocal lattice
- Subdivision the Non Bonded
- reciprocal lattice potential
- The smooth particle mesh
- REDEFINE
- REDEFINE
- reference system
- Potential Subdivision and Multiple
| Subdivision the Non Bonded
- REJECT
- REJECT
- &REM
- &REM
- REM_DIAGNOSTIC
- PRINT_DIAGNOSTIC
- replica exchange method
- Electrostatic Corrections for the
| Serial generalized ensemble simulations
| BATTERIES
| PRINT
| PRINT_ENERGY
| SETUP
- Hamiltonian REM
- Hamiltonian REM
- local scaling and global scaling
- SETUP
- temperature REM
- Temperature REM
- REPLICATE
- REPLICATE
- REPL_RESIDUE
- REPL_RESIDUE
- RESET_CM
- RESET_CM
- RESIDUE
- REPL_RESIDUE
| HBONDS
| RESIDUE
| RESIDUE
- definition of, in the tpg file
- RESIDUE
- sequence
- JOIN
- residue sequence
- DEBUG
- RESTART
- RESTART
| CONTROL
- restart file
- RESTART
| CONTROL
| CHANGE_CELL
- parallel simulation
- RESTART
- restricted canonical transformation
- Canonical Transformation and Symplectic
- reversible integrator
- Liouville Formalism: a Tool
- rigid
- rigid
- root mean square displacement
- HBONDS
| STRUCTURES
| TEMPLATE
| DEF_SOLUTE
- &RUN
- &RUN
- Ryckaert J.-P.
- Equivalence of Atomic and
- SAVE
- SAVE
- SAVE_ALL_FILES
- RESTART
| SETUP
- saving coordinates to disk
- DUMP
- SCALING
- SCALE
| SCALING
- SCALE_CHARGES
- SCALE_CHARGES
- SCALE_MASS
- SCALE_MASS
- scaling
- equivalence of atomic and group
- Equivalence of Atomic and
- SD
- MINIMIZE
- SEGMENT
- SEGMENT
- SEGMENT(&SGE)
- SEGMENT
- SELECT_DIHEDRAL
- SELECT_DIHEDRAL
- Serial Generalized Ensemble simulations
- Serial generalized ensemble simulations
- BAR-SGE method
- The algorithm for optimal
| Implementation of adaptive free
| Free energy evaluation from
- General theory
- Fundamentals of serial generalized-ensemble
- Input of (see also &SGE)
- &SGE
- Simulated tempering
- SGE simulations in temperature-space
- Simulations in collective coordinate space
- SGE simulations in
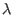 -space
-space
- &SETUP
- &SETUP
- replica exchange method
- SETUP
- SETUP(&SGE)
- SETUP
- &SGE
- &SGE
- FIX_FREE_ENERGY
- FIX_FREE_ENERGY
- PRINT_ACCEPTANCE_RATIO
- PRINT_ACCEPTANCE_RATIO
- PRINT_WHAM
- PRINT_WHAM
- SEGMENT
- SEGMENT
- SETUP
- SETUP
- STEP
- STEP
- TRANSITION_SCHEME
- TRANSITION_SCHEME
- ZERO_FREE_ENERGY
- ZERO_FREE_ENERGY
- Description of the method (see also Serial Generalized Ensemble simulations)
- &SGE
- SHAKE
- Multiple time steps integration
| Constraints and r-RESPA
| CHECK_COORD_OFF
- Shifted potential
- soft-core alternative in alchemical transformations
- SHIFT-POT
- SHIFT-POT
- SHIFT-POT
- Simulated tempering (see also Serial Generalized Ensemble simulations)
- Serial generalized ensemble simulations
- &SIMULATION
- &SIMULATION
- simulation box
- &SETUP
- smooth particle mesh Ewald (see also SPME)
- The smooth particle mesh
- Soft-core Lennard-Jones potential
- Production of the MD
- SOFT-CORE
- SOFT-CORE
- &SOLUTE
- SOLUTE
| &SOLUTE
- defining a fragment of
- &PROPERTIES
| DEF_FRAGMENT
- input examples
- READ_CO
- input topology from ASCII file
- READ_TPG_ASCII
- input topology from tpgprm file
- READ_TPGPRM
- inserting in solvent
- INSERT
- pair correlation function
- GOFR
- setting up the unit cell
- COORDINATES
- thermostatting solute atoms
- THERMOS
- thermostatting solvent atoms
- THERMOS
- topology
- CRYSTAL
- total charge
- DEF_SOLUTE
- solute tempering
- SEGMENT
- &SOLVENT
- SOLVENT
| &SOLVENT
- generating the coordinates
- COORDINATES
| COORDINATES
| GENERATE
- input examples
- SOLUTE
- reading the coordinates of
- ADD_UNITS
| READ_SOLVENT
- setting up the unit cell
- CELL
| COORDINATES
| GENERATE
- space group
- READ_PDB
| COORDINATES
| SPACE_GROUP
- SPACE_GROUP
- SPACE_GROUP
- spherical cutoff
- Subdivision the Non Bonded
- SPME
- Multiple time steps integration
| Subdivision of the ``Bonded''
| The smooth particle mesh
| EWALD
- accuracy
- The smooth particle mesh
- B-spline interpolation
- The smooth particle mesh
- in multiple time scales integrators
- MTS_RESPA
- memory demand
- The smooth particle mesh
- performances
- The smooth particle mesh
- setting work arrays dimensions.
- How to set dimensions
- STOP
- START
| STOP
- steepest descent
- MINIMIZE
- STEER
- STEER
- steered molecular dynamics
- Enhanced sampling in atomistic
| Steered Molecular Dynamics
- adding a time dependent bending
- ADD_STR_BONDS
| ADD_STR_BENDS
- adding a time dependent stretching
- &POTENTIAL
- adding a time dependent torsion
- ADD_STR_COM
- along a curvilinear coordinate
- SOFT-CORE
- printing out the work
- PLOT
- restart
- STEER
- thermal changes
- PLOT
- STEER_PATH
- STEER_PATH
- step
- MTS_RESPA
- STEP(&REM)
- STEP
- STEP(&SGE)
- STEP
- s_test
- MTS_RESPA
- STRESS
- STRESS
- stress tensor
- Multiple Time Steps Algorithms
| ISOSTRESSXY
- STRETCHING
- BOND
- for the solute
- STRETCHING
- printing out
- PRINT_TOPOLOGY
- stretching potential
- Subdivision of the ``Bonded''
| DEBUG
- structure factor
- The smooth particle mesh
| &PROPERTIES
| GOFR
| TIME_CORRELATIONS
- structured commands
- definition of
- Input File.
- VORONOI
- STRUCTURES
| VORONOI
- symplectic
- building integrators
- Liouville Formalism: a Tool
- condition
- Liouville Formalism: a Tool
- condition for canonical transformations
- Canonical Transformation and Symplectic
- integrators
- Enhanced sampling in atomistic
- notation of the equations of motion
- Canonical Transformation and Symplectic
- TEMPERATURE
- TEMPERATURE
- temperature scaling
- with Nosé thermostats
- THERMOS
- TEMPERED
- TEMPERED
- TEMPLATE
- TEMPLATE
- termatom
- termatom
- test-times
- MTS_RESPA
- thermal changes
- STEER
- thermal work
- STEER
- thermalization
- TIME
- THERMOS
- THERMOS
- thermostat
- The Parrinello-Rahman-Nosé Extended Lagrangian
| THERMOS
- Andersen
- ANDERSEN
- Bussi
- BUSSI
- Nosé-Hoover
- THERMOS
- TIME
- TIME
- time dependent bending
- ADD_STR_BONDS
| ADD_STR_BENDS
- time dependent stretching
- &POTENTIAL
- time dependent torsion
- ADD_STR_COM
- TIME_CORRELATIONS
- TIME_CORRELATIONS
- TIMESTEP
- TIMESTEP
- topology
- REPL_RESIDUE
| Input to ORAC : Force
| Topology
- adding extra topology
- ADD_TPG SOLUTE
- from ASCII file
- READ_TPG_ASCII
- from tpgprm file
- READ_TPGPRM
- printing
- PRINT_TOPOLOGY
- torsion
- definition of dihedral angle
- Topology
- TORSION IMPROPER
- TORSION IMPROPER
| imphd
- printing out
- PRINT_TOPOLOGY
- proper
- TORSION PROPER
- torsional potential
- Applications
| Subdivision of the ``Bonded''
| DEBUG
- TORSION IMPROPER
- TORSION IMPROPER
- TORSION PROPER
- TORSION PROPER
| TORSION PROPER
- total
- HBONDS
- TRAJECTORY
- TRAJECTORY
- trajectory file
- &ANALYSIS
| START
| DUMP
| DUMP
| &PROPERTIES
- auxiliary file
- DUMP
- TRANSITION_SCHEME(&SGE)
- TRANSITION_SCHEME
- Trotter formula
- Liouville Formalism: a Tool
- Tuckerman M.
- Multiple time steps integration
- unit cell
- COORDINATES
| CELL
| COORDINATES
| GENERATE
- replicating along selected directions
- READ_PDB
| COORDINATES
| COORDINATES
- unitary transformation
- Liouville Formalism: a Tool
- UPDATE
- UPDATE
- use_neighbor
- GOFR
- use_neighbors
- for hydrogen bonds
- HBONDS
- vacf
- TIME_CORRELATIONS
- valine
- RESIDUE
- velocity
- rescaling
- SCALE
- velocity autocorrelation function
- &PROPERTIES
| TIME_CORRELATIONS
- velocity Verlet
- Liouville Formalism: a Tool
- Verlet
- neighbor list
- VERLET_LIST
- VERLET_LIST
- VERLET_LIST
- very_cold_start
- MTS_RESPA
- virtual variables
- The Parrinello-Rahman-Nosé Hamiltonian and
- Volume calculation
- VORONOI
- Voronoi
- Group Scaling and Molecular
- Voronoi Polihedra
- VORONOI
- Wang-Landau algorithm
- Metadynamics Simulation: history-dependent algorithms
- water
- Group Scaling and Molecular
- properties of
- Electrostatic Corrections for the
- work
- in a SMD simulation
- PLOT
- in alchemical transformation
- Production of the MD
- write
- DUMP
- WRITE_GRADIENT
- MINIMIZE
- WRITE_GYR
- WRITE_GYR
- WRITE_PRESSURE
- WRITE_PRESSURE
- WRITE_TPGPRM_BIN
- WRITE_TPGPRM_BIN
- WTEMPERED
- WTEMPERED
- Xmol animation
- DEF_FRAGMENT
- X_RMS
- DEF_SOLUTE
- xyz format
- PLOT
- ZERO_FREE_ENERGY(&SGE)
- ZERO_FREE_ENERGY
procacci
2021-12-29