Steered Molecular Dynamics
Steered molecular dynamics simulation (SMD) is a technique
mimicking the principle of the atomic force microscopy (AFM). In
practice, one applies a time dependent mechanical external potential
that obliges the system to perform some prescribed motion in a
prescribed simulation time. SMD has been widely used to explore the
mechanical functions of biomolecules such as ligand receptor
binding/unbinding and elasticity of muscle proteins during stretching
at the atomic level[145]. The SMD has also been used in
the past to approximately estimate the potential of mean force
(PMF)8.1
along a given mechanical coordinate (for example a distance or an
angle). The model upon which this technique for estimating the PMF relies
was based on the assumption that the driven motion along the
reaction coordinate 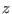 could be described by an over-damped
one-dimensional Langevin equation of the kind
could be described by an over-damped
one-dimensional Langevin equation of the kind
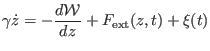 |
(8.1) |
where 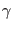 is the friction coefficient,
is the friction coefficient, 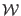 is the underlying
potential of mean force ,
is the underlying
potential of mean force ,
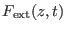 is the external force due
the driving potential and
is the external force due
the driving potential and 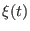 is a stochastic force related to
the friction through the second fluctuation dissipation theorem. The
PMF
is a stochastic force related to
the friction through the second fluctuation dissipation theorem. The
PMF 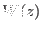 can then be determined only if one knows (or can somehow figure it
out) the friction coefficient, so as to evaluate the frictional force that
discounts the irreversible work done in the driven process. The method
also relies on the strong assumption that the friction along
can then be determined only if one knows (or can somehow figure it
out) the friction coefficient, so as to evaluate the frictional force that
discounts the irreversible work done in the driven process. The method
also relies on the strong assumption that the friction along 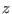 is
local in time, i.e. the underlying equilibrium process is Markovian.
is
local in time, i.e. the underlying equilibrium process is Markovian.
Subsections
procacci
2021-12-29
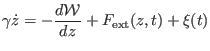
![]() is the friction coefficient,
is the friction coefficient, ![]() is the underlying
potential of mean force ,
is the underlying
potential of mean force ,
![]() is the external force due
the driving potential and
is the external force due
the driving potential and ![]() is a stochastic force related to
the friction through the second fluctuation dissipation theorem. The
PMF
is a stochastic force related to
the friction through the second fluctuation dissipation theorem. The
PMF ![]() can then be determined only if one knows (or can somehow figure it
out) the friction coefficient, so as to evaluate the frictional force that
discounts the irreversible work done in the driven process. The method
also relies on the strong assumption that the friction along
can then be determined only if one knows (or can somehow figure it
out) the friction coefficient, so as to evaluate the frictional force that
discounts the irreversible work done in the driven process. The method
also relies on the strong assumption that the friction along ![]() is
local in time, i.e. the underlying equilibrium process is Markovian.
is
local in time, i.e. the underlying equilibrium process is Markovian.