DESCRIPTION
The SETUP command is used to define the lowest scaling factor(s)
(i.e the highest temperature) of the last replica. The number of
replicas in the REMD simulations are equal to the number of processors
passed to the MPI routines (nprocs). The spacing between the replicas
can be defined in the REM.set user-file by setting the irest
integer equal to 2. If only the 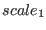 real parameter is specified,
an equal scaling is applied to all parts of the potential. If the
three parameters
real parameter is specified,
an equal scaling is applied to all parts of the potential. If the
three parameters 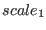 ,
, 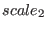 ,
, 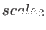 are specified, then
are specified, then
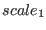 refers to the bending, stretching and improper torsional
potential,
refers to the bending, stretching and improper torsional
potential, 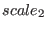 to the (proper) torsional potential and to the 14
non-bonded interactions and finally
to the (proper) torsional potential and to the 14
non-bonded interactions and finally 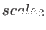 refers to the non bonded
potential.NB: when the Ewald summation is used together with the
command SEGMENT(&REM),
refers to the non bonded
potential.NB: when the Ewald summation is used together with the
command SEGMENT(&REM), 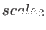 scales only the direct
(short-ranged) part of the electrostatic interactions and the
(long-ranged) reciprocal part has a scaling factor of 1.0 (i.e. these
interaction are not scaled).
If irest=0, the run is restarted from a previous one. This
implies that the directories PARXXXX are present and are equal in
number to nprocs as specified in the mpiexec/mpirun
command. The default prefix of the REM directories can be changed from PAR to anything by specifying in the preamble of the main input file the
shebang directive
scales only the direct
(short-ranged) part of the electrostatic interactions and the
(long-ranged) reciprocal part has a scaling factor of 1.0 (i.e. these
interaction are not scaled).
If irest=0, the run is restarted from a previous one. This
implies that the directories PARXXXX are present and are equal in
number to nprocs as specified in the mpiexec/mpirun
command. The default prefix of the REM directories can be changed from PAR to anything by specifying in the preamble of the main input file the
shebang directive
#!&MYPREFIX
If irest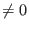 then
the run refers to a cold start from scratch and
then
the run refers to a cold start from scratch and
- if irest
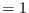 , then the scaling factors of the intermediate
replicas are derived according to a geometric progression, namely
, then the scaling factors of the intermediate
replicas are derived according to a geometric progression, namely
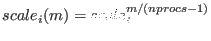 , where
, where
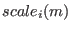 is the
scaling factor for the potential
is the
scaling factor for the potential 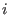 of the replica
of the replica 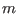 with
with
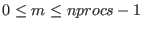 . For example, if
. For example, if
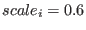 and nprocs=4,
then replica
and nprocs=4,
then replica 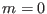 has
has
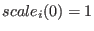 , replica
, replica 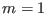 has
has
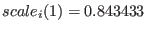 , replica
, replica 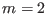 has
has
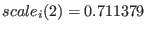 and the replica
and the replica
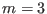 has
has
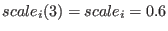 .
.
- if irest
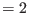 , the scaling factors are read from an
auxiliary file called ``REM.set'' that must be present in the
directory from which the program is launched using the
mpiexec/mpirun command.
This ASCII file has as many lines as parallel processes and on each line
the three (or one) scale factors must be specified.
, the scaling factors are read from an
auxiliary file called ``REM.set'' that must be present in the
directory from which the program is launched using the
mpiexec/mpirun command.
This ASCII file has as many lines as parallel processes and on each line
the three (or one) scale factors must be specified.
- if irest
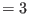 , then each replica read from a user-defined
pdb file that must reside in the PARxxx dir. This is useful when
restarting a REM simulation with the SAVE_ALL_FILES
directive. To restart a REM
with the SAVE_ALL_FILES directive on, the user must hence do
the following: 1) set irest to 3 in the main rem input; 2)
change the filename specification in the SAVE_ALL_FILES
directive (namelist &INOUT) ; this avoids overwriting the saved
restart files ; 3) copy the last configuration in the PARxxx pdb files
to a common user-name file in the same directories (e.g. RESTART.pdb) and instructs the program to read from that PDB in the
namelist &SETUP using the directive READ_PDBb
RESTART.pdb with no ../ specification ;4) optionally rename in
the main input the output files to be produced during the rem
simulation.
, then each replica read from a user-defined
pdb file that must reside in the PARxxx dir. This is useful when
restarting a REM simulation with the SAVE_ALL_FILES
directive. To restart a REM
with the SAVE_ALL_FILES directive on, the user must hence do
the following: 1) set irest to 3 in the main rem input; 2)
change the filename specification in the SAVE_ALL_FILES
directive (namelist &INOUT) ; this avoids overwriting the saved
restart files ; 3) copy the last configuration in the PARxxx pdb files
to a common user-name file in the same directories (e.g. RESTART.pdb) and instructs the program to read from that PDB in the
namelist &SETUP using the directive READ_PDBb
RESTART.pdb with no ../ specification ;4) optionally rename in
the main input the output files to be produced during the rem
simulation.
![]() then
the run refers to a cold start from scratch and
then
the run refers to a cold start from scratch and
![]()