EXAMPLES
&SETUP
CRYSTAL 20.00 20.00 20.00 90.0 90.0 90.0
# REPLICATE 2 2 2
&END
&SOLUTE
COORDINATES solute.pdb
# SPACE_GROUP OPEN benz.group P 2/c
&END
&SOLVENT
CELL SC
INSERT 1.5
COORDINATES solvent.pdb
GENERATE RANDOMIZE 4 4 4
# GENERATE RANDOMIZE 8 8 8
&END
In this example the coordinates of the solute are read in form the
file solute.pdb while the coordinates of the solvent molecule
(see &SOLVENT) are read in form the file solvent.pdb.
As is now, this input would produce in a box of
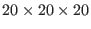 Å
Å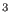 , 1 solute along with 64 replicas (see command
GENERATE(&SOLVENT) of the solvent molecule. Of this 64
molecule, those that overlap with the solute molecule (see command INSERT(&SOLVENT) ) are discarded.
If the second line in the environment &SOLUTE is uncommented,
the solute is assumed to be arranged in the MD box according to the
space group specified by the SPACE_GROUP directive. In the
present example the group contains 4 molecules per unit cell.
So 4 molecules of solute are arranged in the box according to
the P 2/c space group along with 64 replicas of solvent molecules.
Again the overlapping solvent molecules (say
, 1 solute along with 64 replicas (see command
GENERATE(&SOLVENT) of the solvent molecule. Of this 64
molecule, those that overlap with the solute molecule (see command INSERT(&SOLVENT) ) are discarded.
If the second line in the environment &SOLUTE is uncommented,
the solute is assumed to be arranged in the MD box according to the
space group specified by the SPACE_GROUP directive. In the
present example the group contains 4 molecules per unit cell.
So 4 molecules of solute are arranged in the box according to
the P 2/c space group along with 64 replicas of solvent molecules.
Again the overlapping solvent molecules (say 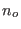 ) will be discarded. If we
comment the line GENERATE RANDOMIZE 4 4 4 and uncomment
the lines # GENERATE RANDOMIZE 8 8 8 and # REPLICATE 2 2 2
we double the size of the sample: we will have 8 cell of
) will be discarded. If we
comment the line GENERATE RANDOMIZE 4 4 4 and uncomment
the lines # GENERATE RANDOMIZE 8 8 8 and # REPLICATE 2 2 2
we double the size of the sample: we will have 8 cell of
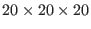 Å
Å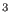 each with 4 molecules of solute and
each with 4 molecules of solute and
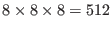 solvent molecules minus
solvent molecules minus
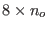 overlapping
molecules.
overlapping
molecules.
![]()