We have seen in section 3.3 that the center of mass or
molecular pressure is equivalent to the atomic pressure. The
atomic pressure is the natural quantity that enters in the virial
theorem [12] irrespective of the form of the interaction
potential among the particles. So, in principle it is safer to adopt
atomic scaling in the extended system constant pressure simulation.
For systems in confined regions, the equivalence between atomic or
true pressure and molecular pressure (see sec. 3.3)
holds for any definition of the molecular subsystem
irrespective of the interaction potentials. In other words we could
have defined virtual molecules made up of atoms selected on different
real molecules. We may expect that, as long as the
system, no matter how its unities or particles are defined,
contains a sufficiently large number of particles, generates a
distribution function identical to that generated by using the
``correct'' atomic scaling.
From a computational standpoint molecular scaling is superior to
atomic scaling. The fast varying Liouvillean in
Eq. (3.70) for the atomic scaling contains the two terms
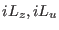 . These terms are slowly varying when molecular
scaling is adopted and are assigned to the slow part of the
Liouvillean in Eq. (3.69). The inner part of the time
propagation is therefore expected to be more expensive for the
multiple time step integration with atomic scaling rather than with
molecular scaling. Generally speaking, given the equivalence
between the molecular and atomic pressure, molecular scaling should be
the preferred choice for maximum efficiency in the multiple time step
integration.3.10
. These terms are slowly varying when molecular
scaling is adopted and are assigned to the slow part of the
Liouvillean in Eq. (3.69). The inner part of the time
propagation is therefore expected to be more expensive for the
multiple time step integration with atomic scaling rather than with
molecular scaling. Generally speaking, given the equivalence
between the molecular and atomic pressure, molecular scaling should be
the preferred choice for maximum efficiency in the multiple time step
integration.3.10
For large size molecules, such as proteins, molecular scaling might be
inappropriate. The size of the molecule clearly restricts the number of
particles in the MD simulation box, thereby reducing the
statistics on the instantaneous calculated molecular pressure which
may show nonphysical large fluctuations. Group
scaling [27] is particularly convenient for handling the
simulation of macromolecules. A statistically significant number of
groups can be selected in order to avoid all problems related to
the poor statistics on molecular pressure calculation for samples
containing a small number of large size particles. Notwithstanding,
for solvated
biomolecules and provided that enough solvent molecules are included,
molecular scaling again yields reliable results. In
Ref. [27] Marchi and Procacci showed that the scaling method in
the
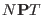 ensemble does not affect neither the equilibrium
structural and dynamical properties nor the kinetic of non equilibrium
MD. For group-based and molecular-based scaling
methods in a system of one single molecule of BPTI embedded in a box
of about a 1000 water molecules, they obtained identical results for
the system volume, the Voronoi volumes of the proteins and for
the mean square displacement of both solvent and protein atoms under
normal and high pressure.
ensemble does not affect neither the equilibrium
structural and dynamical properties nor the kinetic of non equilibrium
MD. For group-based and molecular-based scaling
methods in a system of one single molecule of BPTI embedded in a box
of about a 1000 water molecules, they obtained identical results for
the system volume, the Voronoi volumes of the proteins and for
the mean square displacement of both solvent and protein atoms under
normal and high pressure.
procacci
2021-12-29
![]() ensemble does not affect neither the equilibrium
structural and dynamical properties nor the kinetic of non equilibrium
MD. For group-based and molecular-based scaling
methods in a system of one single molecule of BPTI embedded in a box
of about a 1000 water molecules, they obtained identical results for
the system volume, the Voronoi volumes of the proteins and for
the mean square displacement of both solvent and protein atoms under
normal and high pressure.
ensemble does not affect neither the equilibrium
structural and dynamical properties nor the kinetic of non equilibrium
MD. For group-based and molecular-based scaling
methods in a system of one single molecule of BPTI embedded in a box
of about a 1000 water molecules, they obtained identical results for
the system volume, the Voronoi volumes of the proteins and for
the mean square displacement of both solvent and protein atoms under
normal and high pressure.