DESCRIPTION
This command can be used to impose an additional stretching constraint
between atom iat1 and iat2 of the solute. The numeric
order of the solute atom indices iat1, iat2 is that specified in the topology
file (see 10.3). The added stretching potential has force
constant k (in Kcal/mol/Å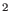 ) and equilibrium distance
) and equilibrium distance
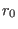 (in Å). If
(in Å). If 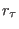 is also specified, then the added stretching
potential is time dependent and
is also specified, then the added stretching
potential is time dependent and 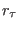 is the equilibrium distance
after the steering time
is the equilibrium distance
after the steering time 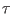 (see STEER(&RUN)) command for the
definition of the steering time in a SMD simulation)
(see STEER(&RUN)) command for the
definition of the steering time in a SMD simulation)
[]WARNINGS
If the chosen 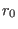 is very different from the actual value of the distance
is very different from the actual value of the distance
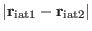 at time 0, a very large force
is experienced by the atoms in involved in the added stretching and the simulation may
catastrophically diverge after few steps.
[]EXAMPLES
at time 0, a very large force
is experienced by the atoms in involved in the added stretching and the simulation may
catastrophically diverge after few steps.
[]EXAMPLES
- Example 1.
ADD_STR_BONDS 1 104 400. 31.5
- Example 2.
&PARAMETERS
READ_TPGPRM_BIN ala10_A.prmtpg
&END
...
&POTENTIAL
...
ADD_STR_BONDS 1 104 400. 31.5 15.5
..
&END
....
...
&RUN
CONTROL 2
...
REJECT 0.0
STEER 10000. 50000.
TIME 50009.0
...
&END
..
&INOUT
RESTART
rmr ../RESTART/ala10 0
&END
In the first example a stretching constraint is imposed between atom 1
and atom 104 of the solute. In the second example a time-dependent
driving potential is applied to the same atoms of the solute. The equilibrium distance
of such harmonic driving potential move at constant velocity in 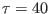 ps
(starting at t=10 ps ) between
ps
(starting at t=10 ps ) between
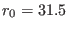 and
and
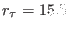 . Since the directive rmr (or
read_multiple_restart ) is
issued in the RESTART(&INOUT) command, the example is assumed to run in
parallel. Each process reads a different restart file named ala10
. Since the directive rmr (or
read_multiple_restart ) is
issued in the RESTART(&INOUT) command, the example is assumed to run in
parallel. Each process reads a different restart file named ala10 .rst in the ../RESTART directory. Note that the
path of the restart files is specified with respect to the actual
value of the pwd command when the parallel version is executed
(i.e. in the
.rst in the ../RESTART directory. Note that the
path of the restart files is specified with respect to the actual
value of the pwd command when the parallel version is executed
(i.e. in the  directories).
directories).
![]() is very different from the actual value of the distance
is very different from the actual value of the distance
![]() at time 0, a very large force
is experienced by the atoms in involved in the added stretching and the simulation may
catastrophically diverge after few steps.
[]EXAMPLES
at time 0, a very large force
is experienced by the atoms in involved in the added stretching and the simulation may
catastrophically diverge after few steps.
[]EXAMPLES
![]()